Executive summary
- The Therapeutic Goods Administration (TGA) Laboratories Branch is responsible for the testing and batch-release of all vaccines supplied in Australia.
- Batches of Pfizer’s Comirnaty® and Moderna’s Spikevax™ COVID-19 vaccines are the two currently available mRNA vaccines in Australia. Comprehensive testing of these vaccines was conducted to ensure they met Australian requirements before distribution.
- A total of 167 batches of the Comirnaty® (n=119) and Spikevax™ (n=48) vaccines that are the subject of this report were supplied in Australia between 9 February 2021 to 26 July 2024. RNA identity, content, integrity, potency, encapsulation, lipid content and identity, lipid nanoparticle size and polydispersity and endotoxin content were assessed. Each batch was tested according to the parameters defined in the manufacturer's batch-release specifications and/or suitable alternative methods. Some of these methods are published in pharmacopoeias such as the European Pharmacopoeia.
- All 167 batches of vaccines supplied in Australia passed the specifications for all quality parameters tested, and there was a high level of comparability between batches with no adverse trends observed.
- Our findings support the decisions made by the TGA and other regulators to authorise these vaccines for use. Batch release of these vaccines will continue according to the TGA Laboratories batch release program.
Background
The severe acute respiratory syndrome-related coronavirus (SARS‑CoV‑2) messenger ribonucleic acid (mRNA) vaccines are a relatively new medical technology. These vaccines contain modified non-replicating mRNA that encode the spike protein of SARS‑CoV‑2 that is enclosed in lipid nanoparticles (LNPs)1. When administered, the target mRNA is introduced inside a patient’s cells, and the sequence of RNA is translated to produce copies of spike proteins that are presented to the immune system outside the host cell wall2-6 (Figure 1). This technology is significant in its ability to: enhance protein functionality while minimising off-target immunogenicity, improving protein folding and assembly, and increasing the efficiency of production and transport of transmembrane and intracellular proteins3. The novel mRNA technology is applicable to both monovalent vaccines, which target a single viral strain, and multivalent vaccines, which are engineered to protect against more than one distinct viral strains, thereby broadening the scope of the immune response.
Figure 1
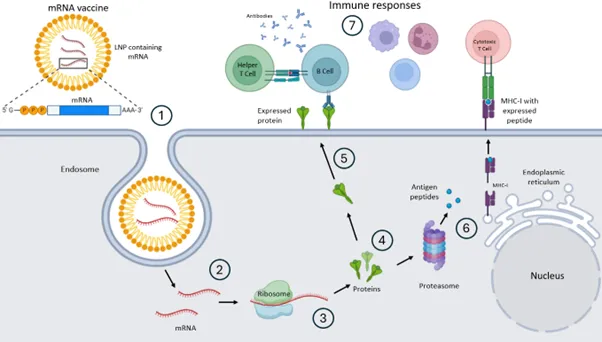
Figure generated using BioRender©.
- mRNA vaccine is delivered into the host cell via endocytosis.
- mRNA is released into the cytosol.
- ribosomes immediately translate constructs to produce the protein of interest.
- The newly formed protein of interest migrates towards the transmembrane.
- The protein is expressed at the cell surface.
- Proteins may be degraded by free proteosomes into peptides that are expressed on major histocompatibility complex (MHC) surface proteins.
- The target protein or expressed peptide is detected by the innate and adaptive immune systems.
In 2021, the TGA provisionally authorised two vaccines that use mRNA technology (Pfizer - Comirnaty® and Moderna - Spikevax™). Both vaccines have since received full registration on the Australian Register of Therapeutic Goods (ARTG) and are now the predominant SARS CoV 2 vaccines supplied in Australia7.
To date, more than 13 billion doses of mRNA vaccines have been administered worldwide. As a result of this widespread use, extensive data on their safety and efficacy in now available (Table 1)8.
In Australia, prescription medicines such as vaccines must meet strict quality, safety and efficacy requirements. The test methods used to verify the quality of each batch must be evaluated and approved by the TGA. These methods must be validated according to International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) Q2(R1) Validation of Analytical Procedures, and the data must be submitted to the TGA for authorisation9. The Sponsor must also submit the proposed range of acceptable results for review by the TGA quality evaluators as part of their submission.
| Study type | Vaccine | Findings | Ref |
|---|---|---|---|
| Meta-analysis | Pfizer BNT162b2 | Nineteen studies demonstrated significant protective effect against confirmed COVID-19 ≥ 14 days after the first dose of 53%, and ≥ 7 days after the second dose interval of 95%. | 10 |
| Meta-analysis | Pfizer BNT162b2, and Moderna mRNA-1273 | A total of 32 human and 23 non-human studies included in this meta-analysis for Pfizer BNT162b2, and Moderna mRNA-1273 achieved >94% efficacy. | 11 |
| Meta-analysis | Pfizer BNT162b2, and Moderna mRNA-1273 | A total of 93 studies were included for BNT162b2, 3 for mRNA-1273 and 26 for both types. The study provided evidence in support for the safety of mRNA vaccines. Most events were mild to moderate, transient and self limiting. | 12 |
| Clinical trial | Moderna mRNA-1273 | The mRNA-1273 vaccine demonstrated a 94.1% vaccine efficacy at preventing symptomatic COVID-19 illness (n=30,420) with no safety concerns identified. | 13 |
| Meta-analysis | Pfizer BNT162b2 | The study included 884,828 participants and defined the risk of developing conditions such as pericarditis after vaccination. The risk for most adverse events was lower than for the same conditions after COVID infection. These findings provide important information regarding vaccine safety and ensure any potential risks are well understood. The TGA has since updated the product information to include these risks. | 14 |
| Meta-analysis | Pfizer BNT162b2, and Moderna mRNA-1273 | A total of 17 studies with 10,935,541 vaccinated and 2,635,251 unvaccinated children aged 5 to 11 years demonstrated a lower risk of SARS-CoV-2 infections, severe COVID-19–related illnesses, and hospitalizations due to COVID-19 following mRNA vaccine administration. | 15 |
| Clinical trial | Pfizer BNT162b2 | A two-dose regimen given 21 days apart demonstrated 95% protection against COVID-19 in 43,548 patients. The incidence of serious adverse events was low and similar between the vaccine and placebo patient groups. | 16 |
| Report | Pfizer-BioNTech/ Comirnaty®, Moderna/Spikevax™ | The Singapore Health Sciences Authority reported a decrease in COVID-19 related adverse events and no new safety signals have arisen with myocarditis recognised as a potential risk following mRNA vaccination at a low of 0.2 incidences per 100,000 doses for bivalent vaccinations and 1.1 per 100,000 doses for monovalent vaccines. | 17 |
Whilst there are a large number of studies that report on the safety and efficacy of SARS-CoV-2 mRNA vaccines, limited studies are available that report on the quality assurance testing of real-world SARS‑CoV‑2 mRNA vaccines.
Full TGA registration has now been granted to the latest strain updates of both Comirnaty® and Spikevax™ products and as of 31 July 2024, more than 158 million doses of Comirnaty® and Spikevax™ have been batch-released by the TGA Laboratories in Australia.
- Comirnaty®: >87 million monovalent and >42 million bivalent.
- Spikevax™: >24 million monovalent and >4.8 million bivalent.
The TGA Laboratories undertake batch-release activities including testing on all batches of mRNA COVID-19 vaccines that have been supplied in Australia as it is a critical part of the regulatory oversight of vaccine quality. The testing is in line with the WHO TRS978 Annex 2 WHO Guidelines for independent lot release of vaccines by regulatory authorities18. This study reports the quality of the Pfizer - Comirnaty® and Moderna - Spikevax™ mRNA vaccines supplied to Australia that are included in the ARTG. The objectives of this study included assessing the composition, identity, potency, and purity of the formulation of these vaccines.
Methods
Samples
Batch-release testing for Comirnaty® and Spikevax™ mRNA COVID-19 vaccines in this report occurred between 9 February 2021 to 26 July 2024 (however, batch release of these products is ongoing). Samples from different batches and formulations of Comirnaty® (Pfizer Australia Pty Ltd) and Spikevax™ multidose vials and pre-filled syringes (Moderna Australia Pty Ltd) were used in this study. Samples were stored in accordance with their approved storage conditions.
Test procedures
Table 2 provides a list of analytical test methods used to test mRNA vaccines as a part of the TGA batch-release19-22. Researchers seeking to perform the same assays should request the complete manufacturer's testing protocols directly from each manufacturer due to the commercial confidentiality of some of the testing protocols.
| Quality attribute | Testing method19-21 | Comirnaty® | Spikevax™ |
|---|---|---|---|
| Identity and composition | RNA identity – Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) | all products (excluding WT/BA.1) | monovalent only (AUST R 370599, 388244) |
| RNA identity/ratio ¬– Reverse-phase ion-pair high-performance liquid chromatography (RPIP-HPLC) | bivalent BA.4.5 only (AUST R 400874) | bivalent only (AUST R 389513, 399553) | |
| Lipid content – HPLC with charged aerosol detection (HPLC-CAD) | all products | all products | |
| Integrity and purity | RNA integrity – capillary gel electrophoresis (CGE) | all products | N/A |
| RNA purity – RPIP-HPLC | N/A | all products | |
| Integrity and composition | RNA encapsulation – Spectrophotometry | all products | all products |
| RNA content – Fluorescence | all products | N/A | |
| RNA content – Anion exchange HPLC (AEX-HPLC) | N/A | all products | |
| LNP size and polydispersity index (PDI) – dynamic light scattering (DLS) | all products | all products | |
| Safety | Endotoxin – Limulus amebocyte lysate (LAL) | all products | all products |
| Potency | In vitro expression (IVE)* | all products | all products |
* This is not a test performed for release of the batch. Testing was performed on a subset of batches after their release.
RNA identity - RT-qPCR
To confirm that the correct RNA species is present in the samples, both Comirnaty® and Spikevax™ vaccines were tested by RT-qPCR amplification using sequence-specific primers and probes. RNA was extracted from samples using the QiAMP RNA Viral Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Reactions were performed using the QuantStudio 3 RT-PCR System (Thermo Fisher Scientific, Massachusetts, U.S.), with a commercially available one-step RT-PCR kit. For each assay, nuclease-free water was used as a negative control, and a drug substance (DS) reference material was used as a positive PCR control. Negative extraction controls were prepared using nuclease-free water subjected to the RNA extraction process. Where applicable, positive extraction controls were prepared using a reference batch of vaccine drug product (DP). All samples and controls were tested using three technical replicates with product-specific thermocycling conditions, including a reverse transcription step followed by multiple cycles of melting and annealing. The mRNA sequence identity was confirmed by sample amplification in all test replicates, with cycle threshold (CT) values below a product-specific cut-off. Assay suitability was determined by control performance: all positive control material replicates were required to exhibit CT values below the cut-off, and all negative control replicates were required to exhibit either no amplification or CT values greater than the cut-off value. A sample was accepted if all three replicates produced the consistent results.
RNA identity/ratio - RPIP-HPLC - Spikevax™
This assay confirms the identities and relative content of the two mRNA species present in the bivalent vaccine formulation. To confirm the identity and ratio of RNA in Spikevax™ bivalent products, samples were tested by RPIP-HPLC using the Waters Acquity H-Class ultra-performance liquid chromatography (UPLC) system (Waters, USA). RNA samples were denatured in a deformulation buffer to disrupt secondary RNA structures using a commercially available kit (Zymo Research, USA). RNA was purified by spin-column with a wash buffer, RNase-free water and centrifugation at 15,000 x G for 1 minute. For digestion, prepared RNA samples and template-guided RNase H digestion mixture were incubated at 65°C for 30 minutes followed by the addition of pre-prepared quench solution and DIEAA solution according to the manufacturer’s instructions. Resulting RNA fragments were separated by RPIP-HPLC using the manufacturer’s recommended gradient conditions with a flow rate of 0.45 mL/min. The separated RNA fragments were detected in-line by UV absorbance at 260 nm and peaks corresponding to known RNA standards were used to determine RNA identities. The RNA ratio for samples was determined by comparing the sample RNA % peak area against the % peak area of RNA product standard constants. Acceptance criteria were set as % interference ≤ 5%, % relative standard deviation (RSD) of reference standard RNA peak area ≤ 5%, % RSD of reference standard RNA peak retention time ≤ 5%, % recovery of each bracketing standard peak area as compared to the average peak area of reference standard RNA peak of the reference standard 90 – 110%.
Lipid content - HPLC-CAD
This assay confirms the correct type and amount of each of the four lipids that comprise the LNP.
For Comirnaty®, identification and quantitation of ALC-0159, cholesterol, ALC-0315 and DSPC were achieved using the Waters Acquity H-Class UPLC system (Waters, USA) with a charged aerosol detector (CAD) (Thermo Fisher Scientific, Massachusetts, U.S). The mobile phase consisted of a combination of aqueous (e.g., water or buffered solutions) and organic solvents to minimise background noise and ensure compatibility with the CAD. Standard stock preparations were made using an alcohol solution and the lipid reference standards, with controls prepared using a dilution of the stock solution based on the manufacturer’s instructions. DP samples were diluted in methanol to ensure lipid concentrations met the ranges specified in the standard curves specified by the manufacturer. Samples were analysed using HPLC-CAD. Sample acceptance criteria were set as % RSD ≤ 10 as well as % relative retention time (RRT) for each lipid according to the acceptance specified by the manufacturer.
For Spikevax™, identification and quantitation of PEG 2000 DMG, cholesterol, SM-102 and DSPC was achieved using the Waters Acquity H-Class UPLC system (Waters, USA) with a CAD detector (Thermo Fisher Scientific, Massachusetts, U.S). The mobile phase consisted of a combination of aqueous (e.g., water or buffered solutions) and organic solvents to minimise background noise and ensure compatibility with the CAD. Standard stock preparations were made using an alcohol solution and the lipid reference standards, with controls prepared using a dilution of the stock solution based on the manufacturer’s instructions. DP samples were diluted in ethanol to ensure lipid concentrations met the ranges specified in the standard curves specified by the manufacturer. Samples were analysed using HPLC-CAD. Sample acceptance criteria were set as % RSD ≤ 10 as well as % RRT for each lipid according to the acceptance criteria specified by the manufacturer.
mRNA content and encapsulation efficiency
For Comirnaty® samples, to confirm the correct content of mRNA is present in each dose, quantitation of total and % encapsulated RNA in LNP was achieved using RiboGreen fluorescence (Thermo Fisher Scientific, Massachusetts, U.S.) with the SpectraMax M5e spectrophotometer system (Molecular Devices, USA). A standard curve was prepared with DS mRNA based on the manufacturer’s instructions using qualified RNA DS reference material. DP samples consisting of encapsulated RNA were compared against a DS standard curve to measure the amount of free RNA (without non-ionic surfactant) and total RNA (with non-ionic surfactant added to release the encapsulated RNA from the LNPs). RiboGreen was added to bind to RNA and generate a fluorescence signal that was measured to determine the amount of free RNA and total RNA. The amount of encapsulated RNA was calculated by subtracting the free RNA content from the total RNA, and the encapsulation efficiency was calculated as the ratio between encapsulated and total RNA, expressed as a percentage. Assay acceptance criteria included a minimum goodness-of-fit for each standard curve and a maximum variability in fluorescence measurements for the replicates of each standard curve dilution.
For Spikevax™ samples, the amount of total mRNA present in the vaccine dose was determined by AEX-HPLC. Free (unencapsulated) mRNA content was measured by a spectrophotometric assay and used to determine the encapsulation efficiency of Spikevax™ vaccine samples in accordance with the manufacturer’s instructions. Encapsulated mRNA was calculated as the difference between total mRNA as measured by AEX-HPLC (described below) and free mRNA content. Encapsulation efficiency was calculated as the ratio between the encapsulated and total mRNA content, expressed as a percentage. The assay involved the complexation of a dye with free mRNA in vaccine test samples and measurement of the associated change of absorbance. Vaccine test samples were prepared according to the manufacturer’s instructions to a constant concentration and mixed with stock solution before absorbance measurement with the M5e spectrophotometer. Baseline absorbance was determined using the stock solution mixed with vaccine formulation buffer without mRNA. The shift in absorbance is proportional to the free RNA content, which was calculated by applying an empirically determined constant relating to absorbance changes and RNA concentration, as provided by the manufacturer.
RNA integrity - capillary gel electrophoresis
RNA integrity of Comirnaty® vaccines was assessed using capillary gel electrophoresis (5300 Fragment Analyzer System, Agilent, California, U.S). According to the manufacturer’s specifications, samples and reference materials were prepared by incubation with a non-ionic surfactant and alcohol solution to release RNA from LNPs. The released RNA was denatured at 70°C for 2 minutes prior to separation by capillary gel electrophoresis. The analysis was performed using the commercially available Standard Sensitivity (SS) RNA kit (DNF-471-1000, Agilent, California, USA). Separation of RNA according to size was achieved by the application of an 8 kV voltage through the capillary array. The inclusion of a fluorescent intercalating dye in the running gel allowed for detection of RNA fragments as they passed before a LED light source linked to a charge-coupled device (CCD) detector. The migration of RNA through each capillary was normalised using an internal marker, and fragment sizes were determined using an RNA ladder. Samples were tested in technical triplicates, with a blank as a negative control and a reference material DP as a positive control. RNA integrity was reported as the % area of the main peak compared to total RNA present in the sample. The system suitability and assay acceptance criteria involved assessing the specificity and range of the RNA ladder and positive control.
RNA purity - RPIP-HPLC
For Spikevax™, the purity of mRNA in monovalent and multivalent final drug product formulations was determined by RPIP-HPLC with UV detection at 260 nm. The method separates mRNA size variants based on their length by gradient elution using ion-pairing agents in the mobile phases using Waters Acquity H-Class UPLC system (Waters, USA). The purity is reported as the relative area % of the main peak, which represents full-length mRNA.
RNA content - AEX-HPLC
For Spikevax™, quantitation of total RNA content for final drug product samples was achieved using AEX- HPLC with UV detection at 260 nm. Agilent 1260 Infinity II HPLC system (Agilent Technologies, USA) was used. The results were reported as the concentration of total mRNA present in the formulations.
LNP size and polydispersity - DLS
Analysis of hydrodynamic particle size and the polydispersity index PDI of Comirnaty® and Spikevax™ vaccines was achieved using DLS with a Malvern Zetasizer Pro Blue (Malvern Panalytical, UK). NIST Traceable Nanosphere Particle Size Standards (Thermo Fisher Scientific, USA) were used as reference standards to ensure accurate particle size measurement. All work was performed in a biosafety cabinet to ensure a dust-free environment. Mean hydrodynamic diameter and PDI were calculated by the instrument software. Reference standards for Comirnaty® and Spikevax™ samples were diluted to the appropriate concentrations before measurement. For both products, system suitability, assay, and sample acceptance criteria were determined by the accuracy and precision of the measurements for the standards and/or samples.
Endotoxin - LAL
The test for bacterial endotoxins was conducted in accordance with the methods specified in the European Pharmacopoeia 2.6.14 and United States Pharmacopoeia <85>.
In vitro expression
A cell-based in vitro expression (IVE) assay was used to confirm cell transfection by the LNPs, and translation of the mRNA sequence into the spike protein. HEK239T (Human embryonic kidney 239T cells) (ATCC, Washington, DC, USA), were incubated with Comirnaty® and Spikevax™ products according to Pfizer’s testing method. For each assay, nuclease-free water and a DP reference material were used as negative and positive controls, respectively. Post incubation, cells were harvested and stained with a cell viability dye, followed by a fixation step. To detect the spike protein, the fixed cells were incubated with an antibody specific to the SARS‑CoV‑2 spike protein, followed by incubation with a secondary antibody conjugated to phycoerythrin. Immunofluorescent cells were detected on a BD FACSCanto II (Becton Dickinson, New Jersey, USA). Assay acceptance was determined by the number of analysed cells, the percentage of viable cells, the precision of the technical replicates, and the results from the controls. Sample acceptance was determined by the percentage of single, viable fluorescent cells.
Monitoring for residual DNA content
Testing commenced in 2023 using samples from the two most recently supplied COVID-19 vaccine strains (bivalent original +BA.4-5 and monovalent XBB1.5) of both Comirnaty® and Spikevax™ products. Testing was conducted in accordance with the standard test procedures for residual deoxyribonucleic acid (DNA) quantitation as described in the European Pharmacopoeia 2.6.35.
Statistical analysis
The mean, along with the standard deviation and percent coefficient of variation [(standard deviation/mean) x 100] was used as the primary method of analysis to evaluate variability within the test results and assess batch-to-batch consistency.
Results
Samples
All 167 vaccine batches were tested, comprising 119 batches of Comirnaty® and 48 batches of Spikevax™ (Table 3). For some tests, the number of batches tested does not directly correspond with the number released for supply in Australia, due to a range of operational and technical factors. Further details are provided in the footnotes for Table 3.
| AUST R | Comirnaty® | Concentration | Batches released (1) | Identity (RT-qPCR) | Identity/ Ratio (RPIP-HPLC) (2) | Lipid content (HPLC-CAD) | Integrity (CGE) | Encapsulation | Content (Fluoresc.) | LNP size & PDI (DLS) | Endotoxin (LAL) | Potency (IVE) (3) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 346290 | Tozinameran (PBS/Sucrose) | 30 µg/0.3 mL | 78 | 78 | - | 75(a) | 77(a) | 78 | 78 | 60(a) | 80(b) | 3 |
| 377110 | Tozinameran (Tris/Sucrose) | 30 µg/0.3 mL | 2 | 2 | - | 2 | 2 | 2 | 2 | 2 | 2 | - |
| 377111 | Tozinameran | 10 µg/0.2 mL | 15 | 15 | - | 15 | 15 | 15 | 15 | 15 | 15 | 6 |
| 393433 | Tozinameran | 3 µg/0.2 mL | 2 | 2 | - | 1(c) | 2 | 2 | 2 | 2 | 2 | - |
| 394890 | Tozinameran/Riltozinameran (Bivalent BA.1) | 15/15 µg/0.3 mL | 5 | (d) | (d) | 5 | 5 | 5 | 5 | 5 | 5 | - |
| 400874 | Tozinameran/Famtozinameran (Bivalent BA.4-5) | 15/15 µg/0.3 mL | 10 | 8(c) | 7(c) | 10 | 10 | 10 | 10 | 10 | 10 | - |
| 419330 | Raxtozinameran (XBB.1.5) | 30 µg/0.3 mL | 5 | 5 | - | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| 419332 | Raxtozinameran (XBB.1.5) | 3 µg/0.2 mL | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | - |
| 419371 | Raxtozinameran (XBB.1.5) | 10 µg/0.3 mL | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AUST R | Spikevax™ | Concentration | Batches released (1) | Identity (RT-qPCR) | Identity/ Ratio (RPIP-HPLC) (2) | Lipid content (HPLC-CAD) | Purity (RPIP-HPLC) | Encapsulation | Content (AEX-HPLC) | LNP size & PDI (DLS) | Endotoxin (LAL) | Potency (IVE) (3) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 370599 | Elasomeran(b) | 0.2 mg/mL | 24 | 43 | - | 39 | 43 | 43 | 43 | 43 | 43 | 11 |
| 388244 | Elasomeran | 0.1 mg/mL | 2 | 2 | - | 2 | 2 | 2 | 2 | 2 | 2 | - |
| 389513 | Elasomeran/Imelasomeran (Bivalent BA.1) | 0.1 mg/mL | 6 | - | 5(c) | 6 | 6 | 6 | 6 | 6 | 6 | - |
| 399553 | Elasomeran/Davesomeran (Bivalent BA.4-5) | 0.1 mg/mL | 10 | - | 9(c) | 10 | 10 | 10 | 10 | 10 | 10 | 3 |
| 418911 | Andusomeran (XBB.1.5) | 0.1 mg/mL | 6 | 6 | - | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
Represents unique numbers of batches, doesn’t include additional shipments of the same batch.
Testing only performed on bivalent vaccines.
Testing was performed on a subset of batches after their release due to the long lead-time in establishing and validating and performing the IVE method.
Testing commenced once the necessary laboratory equipment and reagents became available.
Additional batches underwent tested, although they were not designated for release in the Australian market.
Testing of some batches was not possible due to technical issues with instrumentation or the temporary unavailability of necessary resources. Other testing results and the protocol review demonstrated compliance.
Pfizer used digital droplet PCR (ddPCR), a technology that was not available to the TGA at the time, necessitating the development of an alternative method.
RNA identity - RT-qPCR
For all tested batches (Comirnaty® n=112, Spikevax™ n=51) the target subsequence was sufficiently amplified (based on CT), confirming RNA identity.
RNA identity/ratio - RPIP-HPLC
A total of 14 bivalent (Wuhan and Omicron) Spikevax™ batches were tested. All results are within the test specifications (Figure 2) and the coefficient of variation (CV) for all datasets is <5%. Seven Comirnaty® batches were tested for information purposes only and fall within the criteria of the Spikevax™ samples.
Figure 2
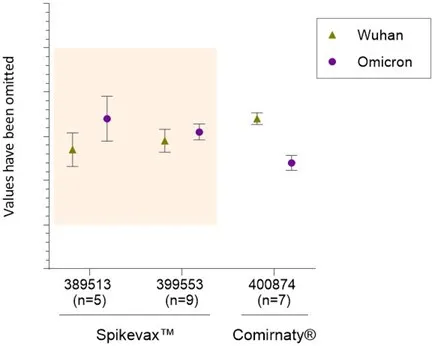
RNA identity and ratio. Data shows the mean and standard deviation for each RNA (Wuhan and Omicron) relative to the assay control. AUST R numbers are given on X-axis, n= the number of batches tested. The y-axis values have been removed to protect commercially sensitive information. Upper and lower specification limits are represented by the coloured box. No limit specifications are shown for Comirnaty®, this data is for information only.
Lipid content - HPLC-CAD
All results were within the test specifications (Figure 3 and Figure 4). For Comirnaty®, all but one of the HPLC-CAD datasets presented in Figure 3 have a CV <10%. The exception is for the ALC-0315 content of AUST R 377110, which has a CV of 11.6%, this is because only 2 batches were received for testing. For Spikevax™, most datasets have a CV ≤12%. The CV values for two datasets are 16.8% and 21.2%, due to all the results being rounded to only one decimal point, unlike other data for this test method.
Figure 3
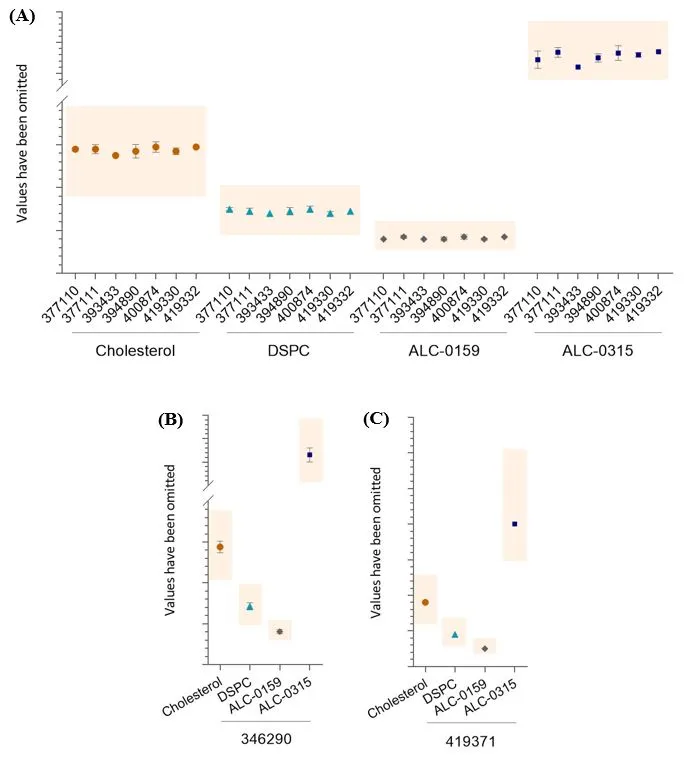
Comirnaty® lipid content. Amount of each lipid measured by HPLC-CAD. Data shows mean and standard deviation. The coloured boxes indicate the upper and lower specification limits.
- AUST R numbers given along the x-axis n= 2, 15, 1, 5, 10, 5, 1, respectively.
- Testing results for AUST R 346290, n= 75.
- Testing results for AUST R 419371, n=1.
The y-axis values have been removed to protect commercially sensitive information. Specifications differ between the different AUST R numbers due to the different formulations of mRNA vaccines (see Table 3).
Figure 4
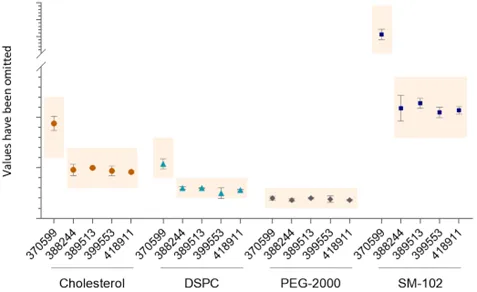
Spikevax™ lipid content. Amount of each lipid measured by HPLC-CAD. Data shows mean and standard deviation. The coloured boxes indicate the upper and lower specification limits. For AUST R numbers given along the x-axis n= 39, 2, 6, 10, 6, respectively. The y-axis values have been removed to protect commercially sensitive information. Specifications differ between the different AUST R numbers due to the different formulations of mRNA vaccines (see Table 3). Note that the individual lipid content may not necessarily align with the mRNA concentration.
RNA integrity - CGE
All results are above the test specification (Figure 5) and the CV for all datasets is <6%.
RNA purity - RPIP-HPLC
All results are above the test specification (Figure 5) and the CV for all datasets is <6%.
Figure 5
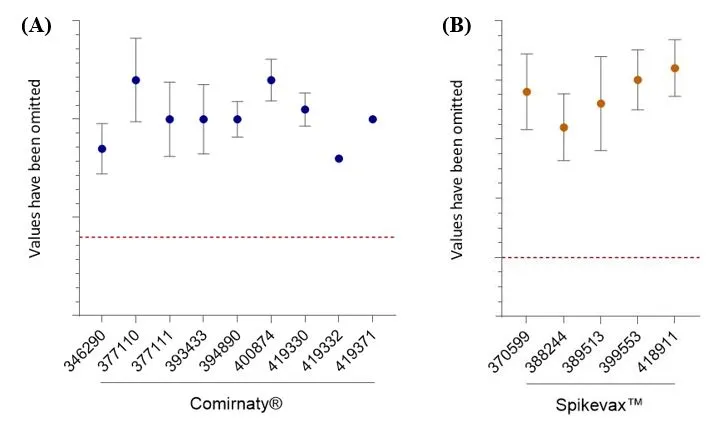
RNA integrity / purity.
- Percentage of full-length transcript as determined by CGE (Comirnaty®) or
- RPIP-HPLC (Spikevax™).
Data shows mean and standard deviation. For Comirnaty® AUST R numbers on the X-axis, n= 77, 2, 15, 2, 5, 10, 5, 1, 1, respectively. For Spikevax™ AUST R numbers on the x-axis, n= 43, 2, 6, 10, 6, respectively. The y-axis values have been removed to protect commercially sensitive information. The lower specification limit (red dashed line) is shown for both Comirnaty® and Spikevax™. RPIP-HPLC and capillary gel electrophoresis yield different results due to their differing analytical principles and sensitivity levels.
RNA encapsulation - spectrophotometry
All results for Comirnaty® are above the test specification (Figure 6) and the CV for all datasets is <3.5%. An anomalous test result was observed during testing of one Spikevax™ bivalent (BA.1) batch, which was later attributed to a testing issue not related to the quality of the batch. Subsequent testing by a European National Control Laboratory (NCL) and retesting by Moderna confirmed the results were above the specification limit and the batch was deemed acceptable.
Figure 6
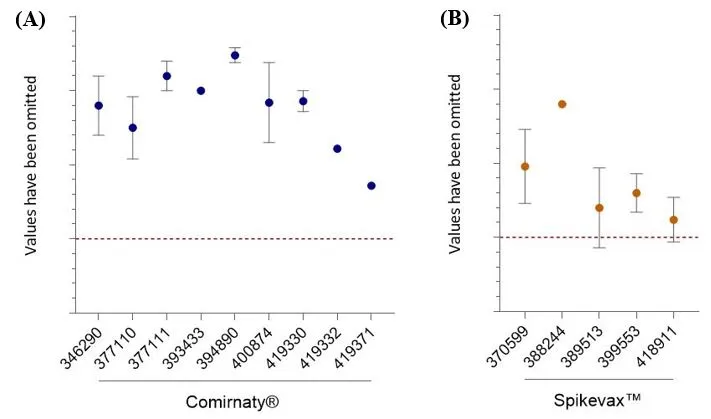
RNA encapsulation. Percentage of encapsulated RNA. Data shows mean and standard deviation. For Comirnaty® AUST R numbers on the X-axis, n= 78, 2, 15, 2, 5, 10, 5, 1, 1, respectively (A). For Spikevax™ AUST R numbers on the x-axis, n= 43, 2, 6, 10, 6, respectively (B). The y-axis values have been removed to protect commercially sensitive information. The lower specification limit is shown as a red dashed line. Note that while the lower error bars for AUST R 389513 and 418911 fall below the specification limit, this is due to the variability in the data and the measurement uncertainty extending below the specification limit and does not indicate maximum and minimum outcomes. All results, excluding one test result for AUST R 389513 were above specification that was later deemed acceptable following additional data.
RNA content - fluorescence
All results are within the test specifications (Figure 7A) and the CV for all datasets is <10.5%.
RNA content - AEX-HPLC
All results have been within the test specifications (Figure 7B). The CV for most datasets is <10%. The one exception is AUST R 388244, which is 14%, however, only two batches of this product have been tested.
Figure 7
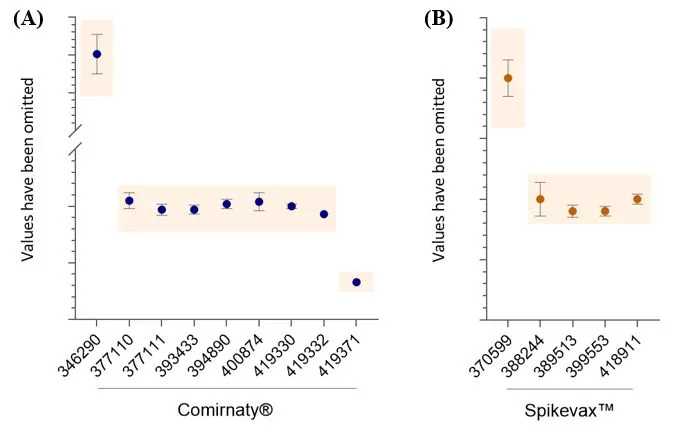
RNA content. Amount of total RNA content (mg/ml) as measured by fluorescence (A) or AEX-HPLC (B). Data shows mean and standard deviation. For Comirnaty® AUST R numbers on the X-axis, n= 78, 2, 15, 2, 5, 10, 5, 1, 1, respectively. For Spikevax™ AUST R numbers on the x-axis, n= 43, 2, 6, 10, 6, respectively. The y-axis values have been removed to protect commercially sensitive information. The upper and lower specification limits are indicated by coloured boxes. Specifications differ between the different AUST R numbers due to the different formulations of mRNA vaccines.
LNP size and polydispersity - dynamic light scattering
All results are within the test specifications (Figure 8). For LNP size, the CV for all datasets is <6.5%. The CV for the PDI is <10 % for most datasets, four are between 10-16%, while for AUST R 393433 the CV is 22%, however only two batches were tested.
Figure 8
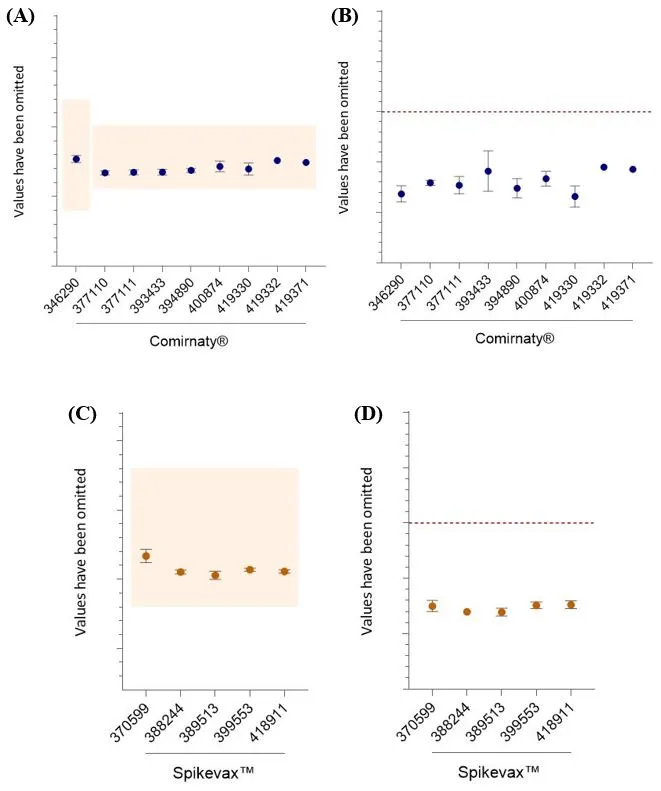
LNP size and polydispersity. Data shows mean and standard deviation for LNP size and polydispersity. For Comirnaty® AUST R numbers on the X-axis, n= 60, 2, 15, 2, 5, 10, 5, 1, 1, respectively. For Spikevax™ AUST R numbers on the x-axis, n= 43, 2, 6, 10, 6, respectively. (A and B) The upper and lower specification limits for size (nm) are indicated by coloured boxes (A and C). The upper limit for the PDI is shown as a red dashed line (B and D).
Endotoxin - LAL
All results were compliant with the test specification. Results are reported as < 5.0 EU/mL (less than 5.0) as the values obtained are below the level of detection of the assay for the dilution used23 (Table 4).
| Vaccine | AUST R | Samples (n=) | Results (EU/mL) |
|---|---|---|---|
| Comirnaty® | 346290 | 80 | < 5.0 |
| 377110 | 2 | < 5.0 | |
| 377111 | 15 | < 5.0 | |
| 393433 | 2 | < 5.0 | |
| 394890 | 5 | < 5.0 | |
| 400874 | 10 | < 5.0 | |
| 419330 | 5 | < 5.0 | |
| 419332 | 1 | < 5.0 | |
| 419371 | 1 | < 5.0 | |
| Spikevax™ | 370599 | 43 | < 5.0 |
| 388244 | 2 | < 5.0 | |
| 389513 | 6 | < 5.0 | |
| 399553 | 10 | < 5.0 | |
| 418911 | 6 | < 5.0 |
In vitro expression
All results were compliant with the test specification for Comirnaty® (Figure 9) and the CV values are all <10.5%.
Figure 9
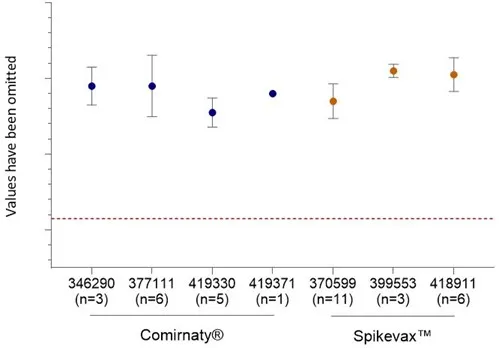
In vitro expression. Percentage of viable, single cells positive for Spike protein. Data shows mean and standard deviation. AUST R numbers are given on X-axis, n= the number of batches tested. The lower specification limit is indicated by the red dashed line.
Monitoring for residual DNA content
All 28 batches tested passed the WHO recommended limit for residual DNA of <10 ng /dose (Table 5)23.
| Vaccine | AUST R | Batches (n=) | Results (ng/dose) | Specification (<10ng/dose) |
|---|---|---|---|---|
| Comirnaty® | 400874 | 8 | 1.59 | <10 |
| 419330 | 6 | 3.85 | <10 | |
| 419332 | 1 | 0.05 | <10 | |
| Spikevax™ | 399553 | 7 | 0.51 | <10 |
| 418911 | 6 | 0.33 | <10 |
Discussion
This study presents an overview of the composition quality of SARS-CoV-2 mRNA vaccines supplied in Australia during the COVID-19 pandemic. All results met approved specifications. For both the Comirnaty® and Spikevax™ mRNA vaccines, no adverse trends were observed across any of the quality assurance parameters. Our results show that where sufficient data analysis is possible, CVs of less than 10% were achieved for the assays, indicating that the variation between the findings is minimal.
The results support the consistency of production across batches supplied for release in Australia and indicate that the manufacturing process for these vaccines is well controlled. Batch-release testing for both Comirnaty® and Spikevax™ mRNA vaccine products, indicate both products are manufactured to a consistently high quality. Our testing should provide the public reassurance that the vaccines comply with established quality requirements, and results show high levels of batch-to-batch consistency. Our testing also confirms the accuracy of the quality tests performed by a manufacturer24. This underscores the robustness of the quality evaluation process for new products before marketing and during post-regulatory authorisation testing by regulatory bodies and NCLs. Recent public interest in levels of residual DNA in SARS‑CoV‑2 mRNA vaccines has prompted the TGA Laboratories to undertake post-release testing of this parameter23, 25. Testing commenced in 2023 using samples from the two most recently supplied SARS‑CoV‑2 vaccine strains (bivalent original +BA.4-5 and monovalent XBB1.5) from both Comirnaty® and Spikevax™, with all test results within the required WHO recommended limit for residual DNA of <10 ng /dose. This testing confirms that both manufacturers minimise the amount of residual DNA fragments in their finished products.
One challenge the scientific community faces is the limited public availability of standardised methods and acceptance criteria for the testing of products manufactured using novel technologies. Sponsors are required to submit detailed proprietary information to regulatory authorities, but such data are not publicly accessible due to their commercial value. The TGA anticipates that standardised, publicly available methods will emerge over time as international standards bodies adapt to evolving technologies.
Conclusion
This study is the largest to date to provide comprehensive evidence supporting the quality assurance, consistency of manufacture and control of Pfizer and Moderna and mRNA vaccines. In total, the TGA released 158 million doses of Comirnaty® and Spikevax™ mRNA vaccines from a total of 167 batches with all batches complying with approved quality requirements and exhibiting high batch-to-batch consistency. Our study supports the reliability of the TGA’s pre-market quality evaluation processes and demonstrates the robustness of the manufacturers’ current quality control measures.
References
- Fiolet T, Kherabi Y, MacDonald CJ, Ghosn J, Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clinical Microbiology and Infection. 2022;28(2):202-21.
- Hermosilla J, Alonso-García A, Salmerón-García A, Cabeza-Barrera J, Medina-Castillo AL, Pérez-Robles R, et al. Analysing the In-Use Stability of mRNA-LNP COVID-19 Vaccines Comirnaty™ (Pfizer) and Spikevax™ (Moderna): A Comparative Study of the Particulate. Vaccines (Basel). 2023;11(11).
- Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nature Biotechnology. 2022;40(6):840-54.
- Mao Q, Xu M, He Q, Li C, Meng S, Wang Y, et al. COVID-19 vaccines: progress and understanding on quality control and evaluation. Signal Transduction and Targeted Therapy. 2021;6(1):199.
- Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nature Reviews Materials. 2021;6(12):1078-94.
- Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nature Reviews Drug Discovery. 2021;20(11):817-38.
- Therapeutic Goods Administration. Batch release assessment of COVID-19 vaccines: Department of Health and Aged Care; 2024 [cited 2 December 2024]. Available from: https://www.tga.gov.au/products/covid-19/covid-19-vaccines/batch-release-assessment-covid-19-vaccines
- International Coalition of Medicines Regulatory Authorities. ICMRA statement on the safety of COVID-19 vaccines 2023 [Available from: https://www.icmra.info/drupal/en/strategicinitiatives/vaccines/safety_statement
- European Medicines Agency. ICH Q2(R2) Validation of analytical procedures - Scientific guideline 2024 [cited 16 December 2024]. Available from: https://www.ema.europa.eu/en/ich-q2r2-validation-analytical-procedures-scientific-guideline
- Kow CS, Hasan SS. Real-world effectiveness of BNT162b2 mRNA vaccine: a meta-analysis of large observational studies. Inflammopharmacology. 2021;29(4):1075-90.
- McDonald I, Murray SM, Reynolds CJ, Altmann DM, Boyton RJ. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. npj Vaccines. 2021;6(1):74.
- Xu W, Ren W, Wu T, Wang Q, Luo M, Yi Y, et al. Real-World Safety of COVID-19 mRNA Vaccines: A Systematic Review and Meta-Analysis. Vaccines. 2023;11(6):1118.
- Baden LR, Sahly HME, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine. 2021;384(5):403-16.
- Barda N, Dagan N, Ben-Shlomo Y, Kepten E, Waxman J, Ohana R, et al. Safety of the BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Setting. New England Journal of Medicine. 2021;385(12):1078-90.
- Watanabe A, Kani R, Iwagami M, Takagi H, Yasuhara J, Kuno T. Assessment of Efficacy and Safety of mRNA COVID-19 Vaccines in Children Aged 5 to 11 Years: A Systematic Review and Meta-analysis. JAMA Pediatrics. 2023;177(4):384-94.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine. 2020;383(27):2603-15.
- Singapore Health Sciences Authority. HSA’s COVID-19 Vaccine Safety Update #15 - 30 December 2020 – 30 June 2023. 2023.
- World Health Organization. Guidelines for Independent Lot Release of Vaccines by Regulatory Authorities Geneva, Switerland2010 [cited 18 November 2024]. Available from: https://cdn.who.int/media/docs/default-source/biologicals/vaccine-quality/guidelines-for-independent-lot-release-of-vaccines-by-regulatory-authorities11a35ea0-f176-4190-948e-8d7d6a7d18d5.pdf?sfvrsn=f19efb94_1
- United States Pharmacopeia. Analytical Procedures for mRNA Vaccine Quality (Draft Guidelines)- 2nd Edition 2023 [Available from: https://www.uspnf.com/notices/analytical-procedures-mrna-vaccines-20230428
- European Medicines Agency. Assessment report COVID-19 Vaccine Moderna 2021.
- European Medicines Agency. Assessment report Comirnaty. 2021.
- Therapeutic Goods Administration. Freedom of Information 4878: Department of Health and Aged Care; 2023 [cited 18 October 2024 2024]. Available from: https://www.tga.gov.au/sites/default/files/2024-06/FOI%204878%20Documents_0.pdf
- Therapeutic Goods Administration. Summary report of residual DNA and endotoxin on CoVID-19 mRNA vaccines conducted by TGA Laboratories: Department of Health and Aged Care; 2024 [cited 16 December 2024]. Available from: https://www.tga.gov.au/resources/publication/tga-laboratory-testing-reports/summary-report-residual-dna-and-endotoxin-covid-19-mrna-vaccines-conducted-tga-laboratories
- Rose NJ, Stickings P, Schepelmann S, Bailey MJA, Burns C. National control laboratory independent lot testing of COVID-19 vaccines: the UK experience. NPJ Vaccines. 2021;6(1):100.
- Therapeutic Goods Administration. Addressing misinformation about excessive DNA in the mRNA vaccines: Department of Health and Aged Care; 2024 [cited 6 December 2024]. Available from: https://www.tga.gov.au/news/media-releases/addressing-misinformation-about-excessive-dna-mrna-vaccines

